Working with CVTDb in R
CvTdb is a database with chemical toxicokinetic data. This post will show you how to use the CvTdb in R with BioBricks.
First, install biobricks:
pip install biobricks
biobricks configure # follow the prompts
Go to docs.biobricks.ai to learn more about the biobricks package.
Then, install the cvtdb asset:
biobricks install cvtdb
biobricks assets cvtdb
# cvtdb_sqlite: .../brick/cvtdb.sqlite
The assets function tells us the path of the sqlite file this asset distributes.
Load the cvtdb sqlite file in R:
library(biobricks)
library(tidyverse)
library(RSQLite)
cvtdb_assets <- biobricks::bbassets("cvtdb")
cvtdb <- dbConnect(RSQLite::SQLite(), cvtdb_assets$cvtdb_sqlite)
RSQLite::dbListTables(cvtdb)
# [1] "chemicals" "conc_time_values" "documents" "series"
# [5] "studies" "subjects" "tk_parameters"
We can see that there are 7 tables in the database.
Click below to see the database schema:
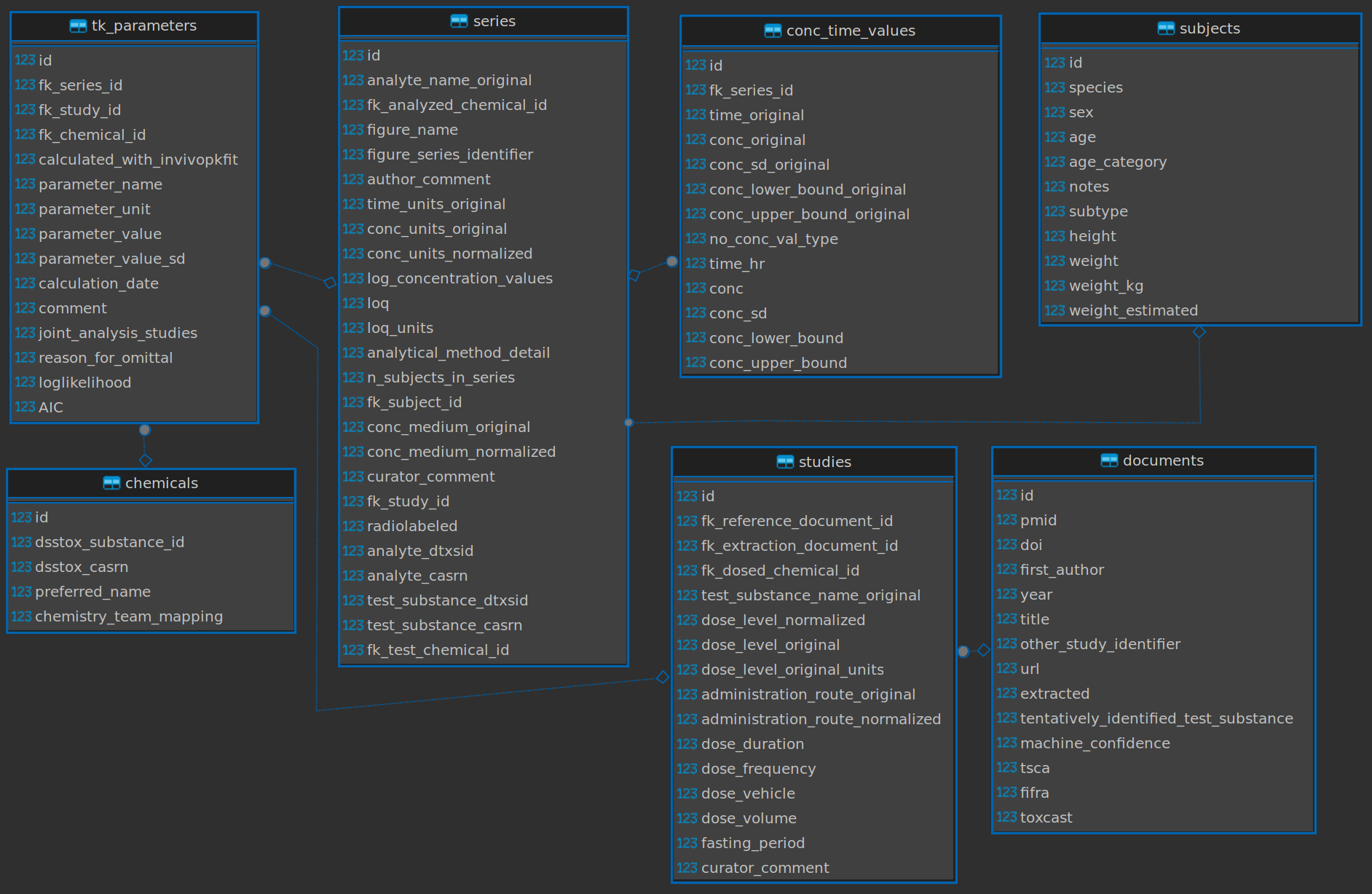
There are 1816 rows in the tk_parameters table which references 756 measured AUC parameters. Let’s make an AUC table that references a chemical, it’s AUC value, and the study from which that came:
chemicals <- tbl(cvtdb, "chemicals") |> collect() |>
select(chemical_id = id, dsstox = dsstox_substance_id)
series <- tbl(cvtdb,"series") |> collect() |>
select(series_id = id, chemical_id = fk_analyzed_chemical_id) |>
inner_join(chemicals,by="chemical_id") |> distinct()
dsstox_auc <- tbl(cvtdb, "tk_parameters") |>
filter(parameter_name=="AUC") |> collect() |>
mutate(value = as.numeric(parameter_value)) |>
inner_join(series,by=c("fk_series_id"="series_id")) |>
select(dsstox,value)
dsstox_auc
# # A tibble: 692 × 2
# dsstox value
# 1 DTXSID3061635 1061.
# 2 DTXSID6044519 1855.
# 3 DTXSID20873970 24276
This is the key data in the cvtdb database. There are a few issues in the cvt database right now (see issue) but I’m sure they will be resolved soon.