Exploring the Diversity of Chemical Reactions in USPTO Data with Python
USPTO Chem_Reaction presents a comprehensive database of chemical reactions compiled from text-mined United States patents issued from 1976 through September 2016. This database is a treasure trove for chemists and data scientists alike, allowing the exploration of chemical reaction data in the detailed Chemical Markup Language (CML) format with the help of BioBricks.
Setting Up BioBricks First off, get BioBricks up and running:
pip install biobricks
biobricks configure # follow the prompts
For more information, refer to the BioBricks documentation.
Installing the Chem_Reaction Asset Proceed by installing the Chem_Reaction asset:
biobricks install Chem_Reaction
Delving into the Data Dive into the dataset with the following Python snippet:
import biobricks as bb
import pyspark
Chem_Reaction = bb.assets('Chem_Reaction')
spark = pyspark.sql.SparkSession.builder.getOrCreate()
# ReactionSmiles PatentNumber ParagraphNum Year TextMinedYield CalculatedYield
# 0 [C:1]([C:5]1[CH:10]=[CH:9][C:8]([OH:11])=[CH:7... US20010000035A1 0007 2001 None None
# 1 [Cl-].[Al+3].[Cl-].[Cl-].[Cl:5][CH2:6][CH2:7][... US20010000038A1 0256 2001 86% 86.9%
# 2 [Al+3].[Cl-].[Cl-].[Cl-].[Cl:5][CH2:6][CH2:7][... US20010000038A1 0259 2001 95% None
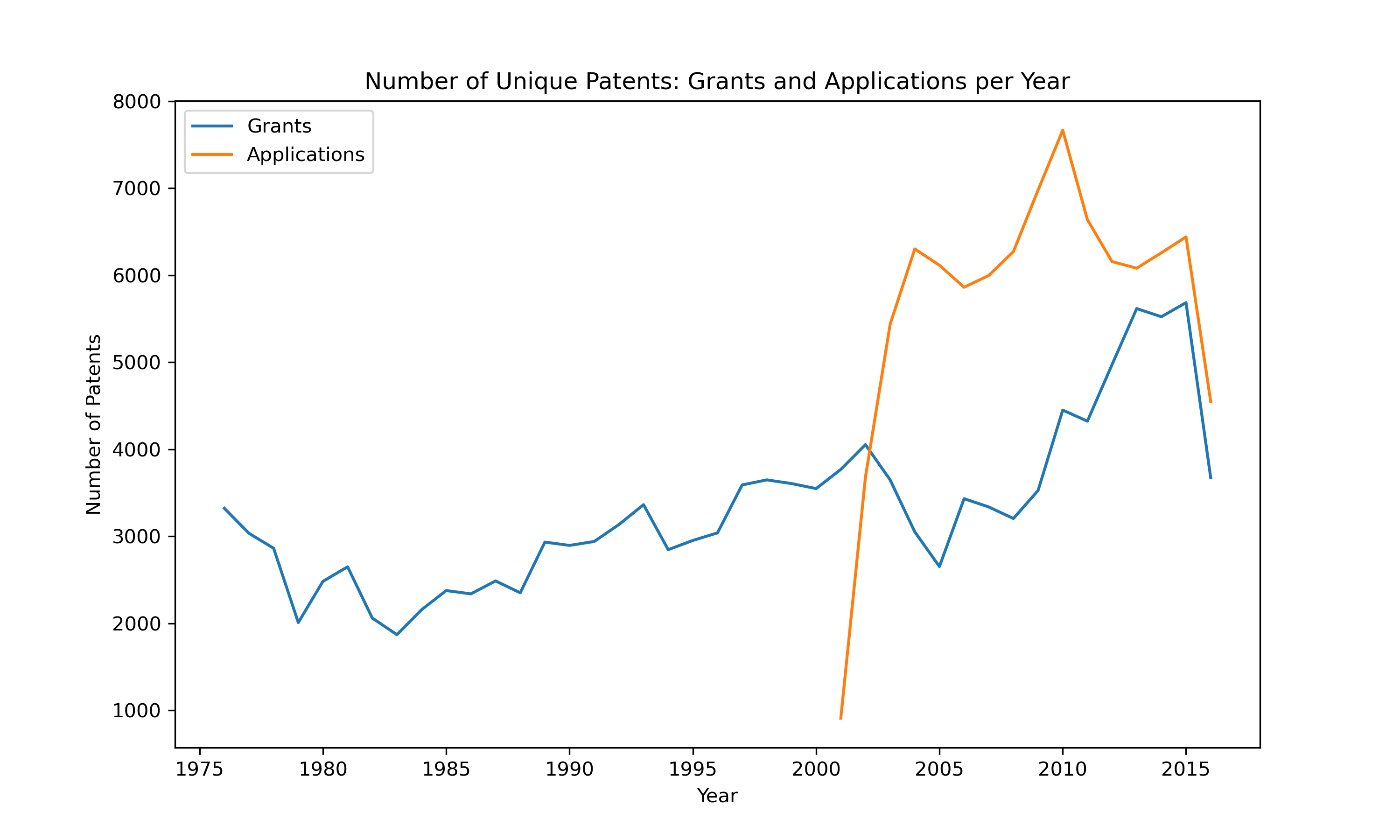
Unveiling the Diversity of Chemicals An intriguing aspect of this dataset is the sheer number of distinct chemical species involved in these reactions. Our analysis revealed:
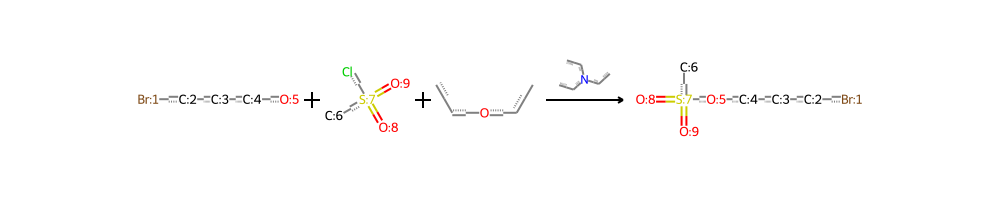
Unique Reactants: 1,216,929 Unique Reagents: 3,520 Unique Products: 1,110,018 These numbers reflect the incredible diversity and complexity of chemical reactions documented in US patents. Additionally, visual representations of these reactions can provide a more intuitive understanding of the chemical processes involved. For instance, consider the following reaction visualization:
This image showcases the transformation of reactants (on the left) into products (on the right), illustrating the intricate details of a chemical reaction at a molecular level.
Additionally, to shed light on the physicochemical properties of the reactants and products involved in these reactions, we have utilized RDKit to calculate molecular weights (MW), partition coefficients (LogP), and the number of rotatable bonds, which are vital descriptors in medicinal chemistry and chemical biology.
reaction_smiles = df['ReactionSmiles'].iloc[1]
reactant_props, product_props = process_reaction_smiles(reaction_smiles)
print("Reactant Properties (MW, LogP, Rotatable Bonds):", reactant_props)
# Reactant Properties (MW, LogP, Rotatable Bonds): [(138.99, 0.764, 2), (114.55, 0.185, 0), (64.04, 0.257, 2)]
print("Product Properties (MW, LogP, Rotatable Bonds):", product_props)
# Product Properties (MW, LogP, Rotatable Bonds): [(217.08, 0.748, 4)]
By examining the atom counts before and after the reaction, we can also infer the type of chemical reaction. In this case, the reaction is identified as an Elimination Reaction, where the number of atoms in the reactants is greater than in the products, indicating the loss of atoms or groups in the reaction. The integration of chemical informatics tools like RDKit in our analysis pipeline not only enhances our understanding of chemical data but also streamlines the process of chemical research, from reaction classification to property prediction and beyond.