Working with CPDB in python
Coding notebook at Working with CPDB in python - Notebook
CPDB is a database about chemicals and cancer. In this post, we’ll use BioBricks.ai to look up chemicals that cause cancer, and learn some basic toxicology. To start, install biobricks.ai and then install cpdb:
pipx install biobricks
biobricks configure # follow the prompts
biobricks install cpdb
CPDB, the Carcinogenic Potency Database, contains a bunch of tables about chemicals and their carcinogenic potency. We can use PySpark to explore it.
import biobricks as bb, pyspark
cpdb = bb.assets('cpdb')
spark = pyspark.sql.SparkSession.builder.getOrCreate()
for table in cpdb.__dict__.keys():
print(table)
# [1]"ncintp_parquet" "ncntdose_parquet" "tumor_parquet"
# [4]"species_parquet" "route_parquet" "chemname_parquet"
# several more ...
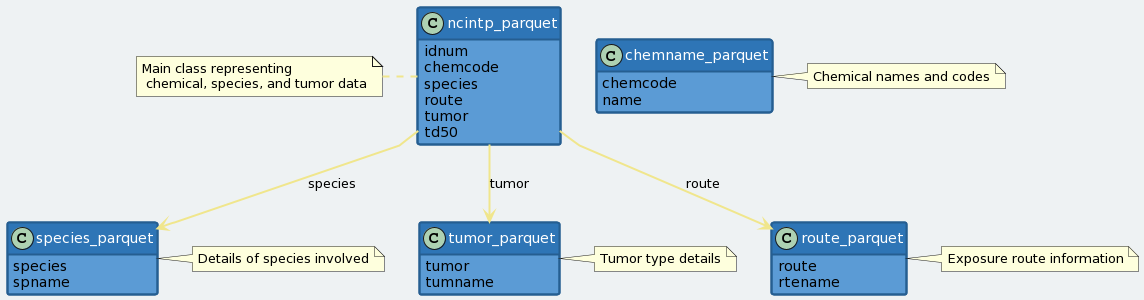
In this analysis we look at 6 tables:
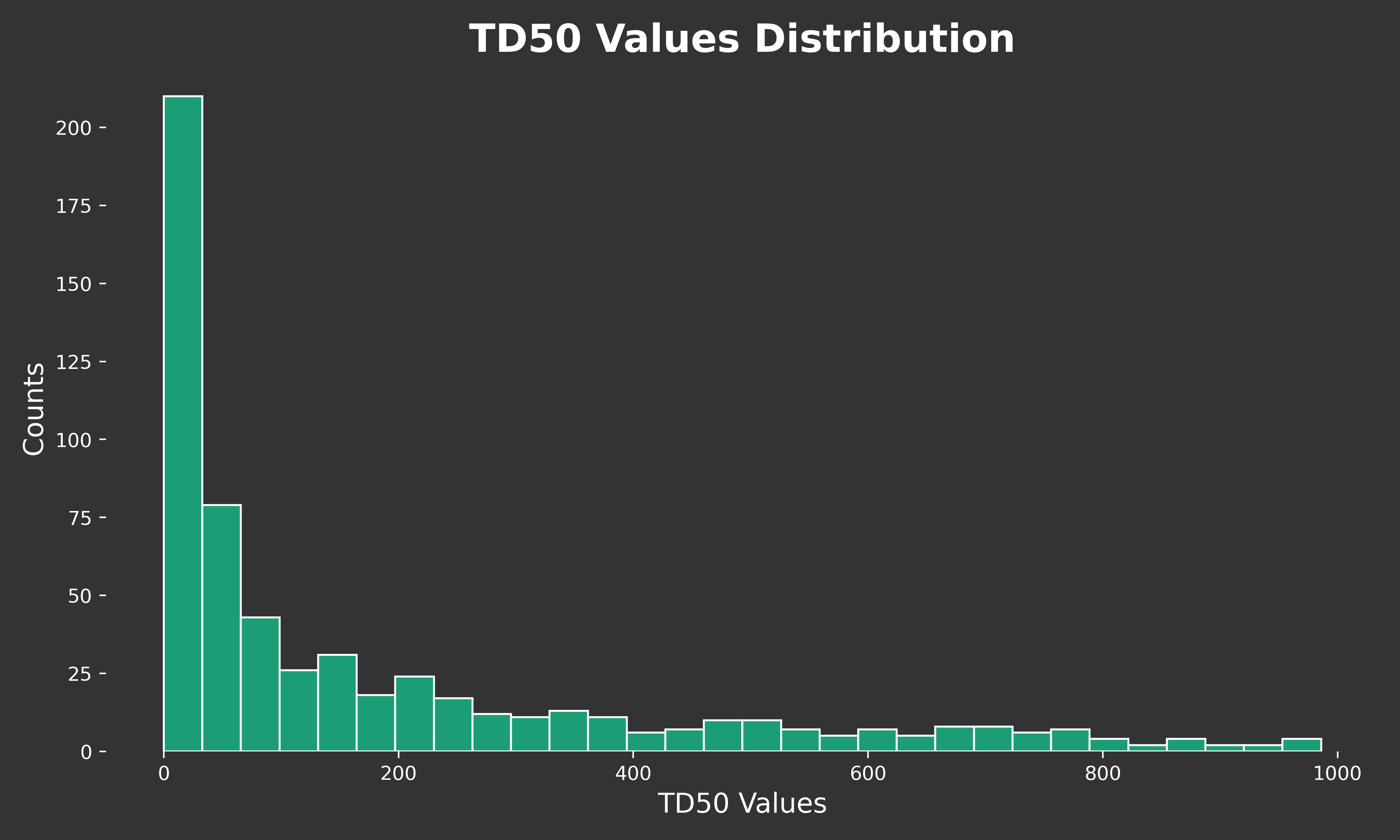
The ncintp_parquet table has a lot of columns, but we can focus on chemcode, species, route and td50. A TD50 describes the dose of a chemical at which 50% of a population will develop cancer. Those doses are measured in mg/kg/day. A TD50 of 1 mg/kg/day means half of a population of 1kg animals given 1mg of a chemical every day will develop cancer.
So let’s write some code to give us an idea of TD50 values for chemicals that cause cancer. We’ll focus on chemicals with a TD50 less than 1000 mg/kg/day. in general, we can consider these compounds carcinogenic.
ncintp = spark.read.parquet(cpdb.ncintp_parquet)
species = spark.read.parquet(cpdb.species_parquet)
route = spark.read.parquet(cpdb.route_parquet)
df = ncintp\
.join(species, 'species')\
.join(route, 'route')\
.select('chemcode','species','spname','route','rtename','td50')
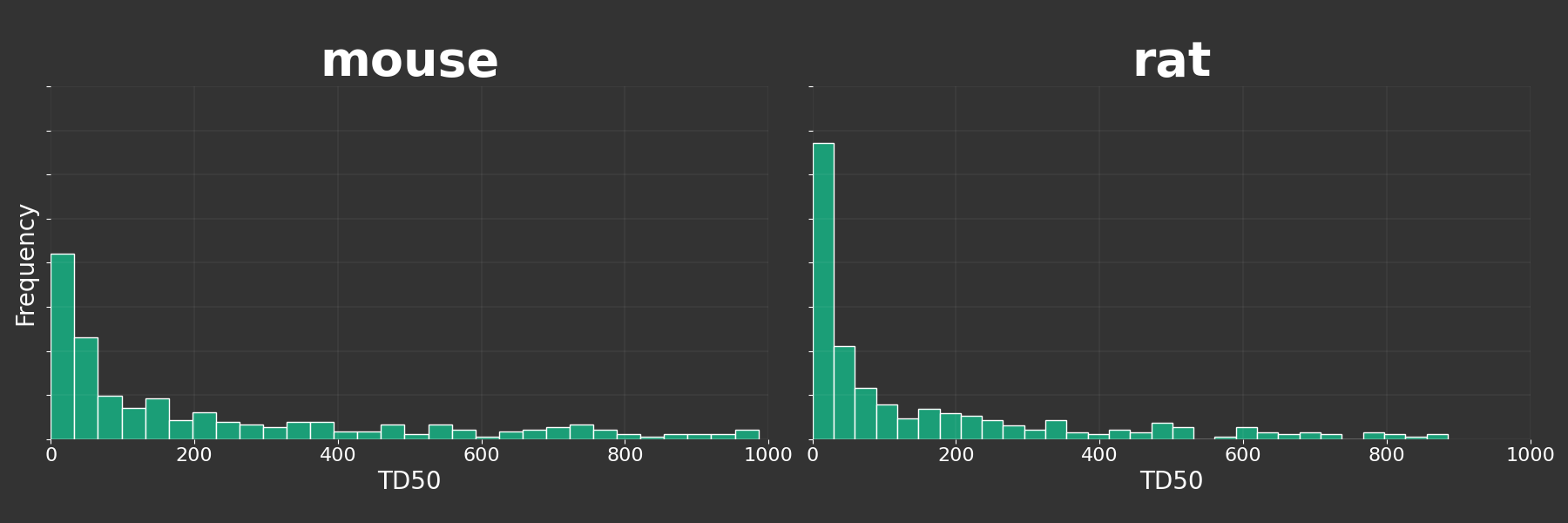
We can build these figures for different species:
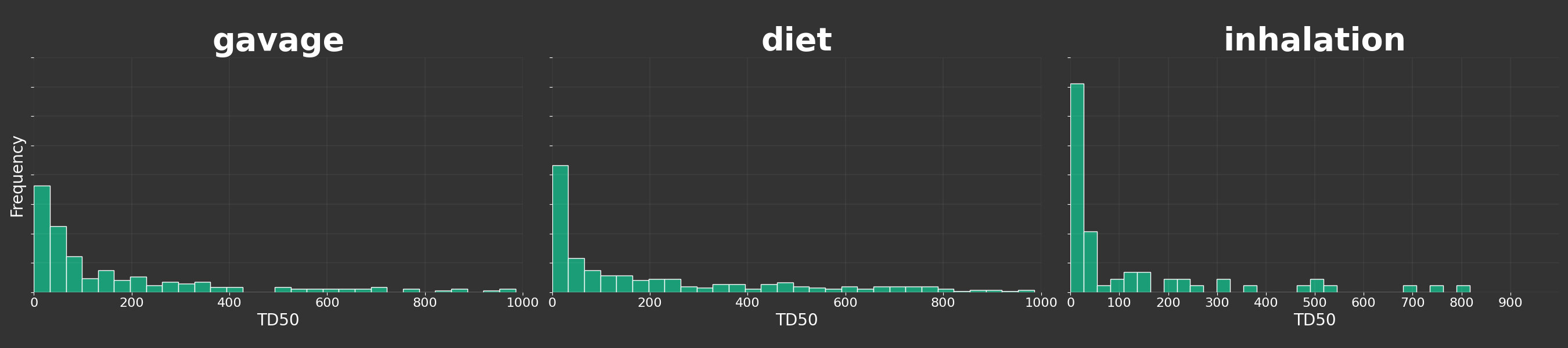
Or for different routes:
Which of these chemicals has the lowest recorded TD50? Let’s take a look:
chemname = spark.read.parquet(cpdb.chemname_parquet)
intp = spark.read.parquet(cpdb.ncintp_parquet)
min_td50 = intp.groupBy('chemcode').agg(F.min('td50').alias('td50'))
df = min_td50.join(chemname,'chemcode').sort(F.col('td50').asc())
df.select('name','td50').show(10,truncate=False)
# +-----------------------------------+-------+
# |name |td50 |
# +------------------------------------+-------+
# |2,3,7,8-TETRACHLORODIBENZO-p-DIOXIN |1.21E-5|
# |HCDD MIXTURE |5.96E-4|
# |o-CHLOROBENZALMALONONITRILE |0.00649|
# |OZONE |0.0156 |
# |RIDDELLIINE |0.0267 |
# |THIO-TEPA |0.0332 |
# |OCHRATOXIN A |0.0579 |
# |POLYBROMINATED BIPHENYL MIXTURE |0.0645 |
# |COBALT SULFATE HEPTAHYDRATE |0.0826 |
# |LASIOCARPINE |0.102 |
# +------------------------------------+-------+
So there you go, by one measure, the most toxic compound in CPDB is 2,3,7,8-TETRACHLORODIBENZO-p-DIOXIN, or TCDD, with a TD50 of 1.21E-5 mg/kg/day. TCDD is a well studied compound, and is known to be highly toxic.